Genetic analyses identified many recurrent mutations in cancer cells and in other disease conditions. By altering or suppressing the function of the corresponding proteins, most of these mutations results in aberrant regulation of cellular biochemical activities. While the functional consequences of some mutations have been characterized in depth, it is often unclear how aberrant regulation of specific biochemical processes affects others. In addition, the functional significance of many observed genetic mutation is still unknown. As a result, the molecular drivers of some cancers and the mechanisms by which cancer cells develop resistance to specific drugging of dysregulated functions (e.g. small molecule inhibition of MAP kinases) are only partially understood.
Direct measurements of protein regulatory events by mass spectrometry proteomics is a powerful approach to probe biochemical regulation in cell homeostasis and in pathological states, such as cancer. The Cifani group develops innovative analytical approaches to measure different modes of biochemical regulation, applying a ‘biological question-driven’ rationale.
Our main areas of interest are:
-
Targeted and ultrasensitive methods to measure differential biochemical regulation and signaling in rare cell populations.
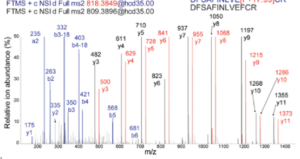
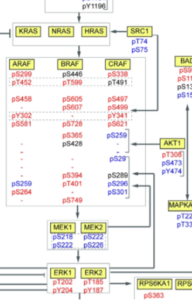
Targeted mass spectrometry coupled to advanced chromatographic methods allows to measure unmodified and chemically modified (e.g. phosphorylated) protein isoforms from minute sample amounts (Cifani & Kentsis, MCP, 2017; Cifani et al, Proteomics, 2017). This led us to develop a panel of targeted peptide assays to probe differential regulation of multiple signaling processes in human cells (QCPA, Cifani & Kentsis, JPR, 2022). We are now building on this rationale to probe regulatory events in rare cell populations (and ultimately in single cells) and to expand it to other regulatory post-translational modification.
-
Rare and non-canonical protein chemical modifications.
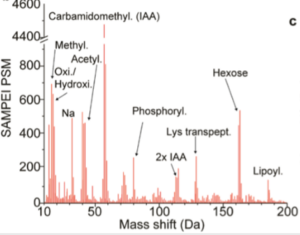
The functional role of phosphorylation and a handful of other PTMs has been extensively investigated, facilitated by the availability of specific detection modes (such as specific antibodies and radioactive probes to study phosphorylation). However, hundreds of different chemical modifications have been observed on mammalian proteins, ranging from different forms of lipidation to transpeptidation, and produced both by enzymatic activities and by reactive chemical species such as metabolic intermediates. What is the full repertoire of chemical modifications in mammalian cells? How many of them have functional roles? How do these rare modifications integrate with known regulatory PTMs? To address these questions, we develop biochemical, chromatographic, mass spectrometric, and computational methods (Cifani et al. JPR, 2021) to discover and measure functional protein modifications.
-
Mapping protein-protein and protein-ligand interactions.
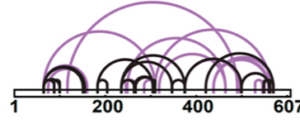
The activity of many proteins is mediated by their physical interaction with other proteins, as in the case of enzymatic complexes, and with other ligands such as cellular macromolecules and drugs. In collaboration with chemists and structural biologists at CSHL, we develop innovative tools to map these interactions with high sequence resolution and sensitivity. To this end we design innovative workflows based on existing reagents for crosslinking (Ser Z, Cifani P, Kentsis A., JPR, 2019) and chemical footprinting, and develop completely new concepts to address biological questions that cannot be addressed with state-of-the-art methods.